4. Analysis frameworks and tools#
4.1. Single-cell analysis frameworks and consortia#
After having obtained the count matrices, as described earlier, the exploratory data analysis phase begins. Due to the size and complexity of the data, specialized tooling is required. While in the early days of single-cell analysis people used to analyze their data with custom scripts, frameworks for specifically this purpose now exist. The three most popular options are the R based Bioconductor[Huber et al., 2015] and Seurat[Hao et al., 2021] ecosystems and the Python based scverse[scverse, 2022] ecosystem. These differ not only in the used programming languages, but also in the underlying data structures and available specialized analysis tools.
Bioconductor is a project which develops, supports and shares free open source software with a focus on rigorous and reproducible analysis of data for many different biological assays including single-cell. A homogeneous developer and user experience and extensive documentation with user friendly vignettes are the biggest strengths of Bioconductor. Seurat is a well regarded R package specifically designed for the analysis of single-cell data. It offers tooling for all steps of the analysis including multimodal and spatial data. The well written vignettes and the large user-base is what Seurat is known for. However, both R options can run into scalability issues for extremely large datasets (more than half a million cells) which motivated the Python based community to develop the scverse ecosystem. scverse is an organization and an ecosystem dedicated to foundational tools in the life sciences with an initial focus on single-cell. Scalability, extendability and strong interoperability with the existing Python data and machine learning tooling are some of the advantages of the scverse ecosystem.
All three ecosystems are involved in many efforts to allow for interoperability of the involved frameworks. This will be discussed in the “Interoperability” chapter. This book always focuses on the best tools for the corresponding question and will therefore use a mix of the above-mentioned ecosystems for specific issues. However, the basis of all analyses will be the scverse ecosystem for two reasons:
While we will regularly switch ecosystems and even programming languages throughout this book, a consistent use of data structures and tooling helps readers to focus on the concepts and not on implementation details.
A great book on exclusively the Bioconductor ecosystem already exists. We encourage users that only want to learn about single-cell analysis with Bioconductor to read it.
In the following sections the scverse ecosystem will be introduced in more detail and the most important concepts will be explained with a focus on the most important data structures. This introduction cannot cover all aspects of the data structures and frameworks. It is not in scope to introduce all available analysis functions. We therefore refer to the respective frameworks’ tutorials and documentation where required.
4.2. Storing unimodal data with AnnData#
As previously discussed, after alignment and gene annotation, genomics data is typically summarized to a feature matrix. This matrix will be of the shape number_observations x number_variables – where for scRNA-seq observations are cellular barcodes and the variables are annotated genes. Over the course of an analysis the observations and variables of this matrix are annotated with computationally derived measurements (e.g. quality control metrics, or latent space embeddings) and prior knowledge (e.g. source donor or alternative gene identifier). In the scverse ecosystem, AnnData[Virshup et al., 2021] is used to associate the data matrix with these annotations. To allow for fast and memory efficient transformations, AnnData also supports sparse matrices and partial reading.
While AnnData is broadly similar to data structures from the R ecosystems (e.g. Bioconductor’s SummarizedExperiment or Seurat’s object), R packages use a transposed feature matrix.
At its core, an AnnData object stores a sparse or dense matrix (the count matrix in the case of scRNA-Seq) in X. This matrix has the dimensions of obs_names x var_names where the obs (=observations) correspond to the cells’ barcodes and the var (=variables) correspond to the gene identifiers. This matrix X is surrounded by Pandas DataFrames obs and var which save annotations of cells and genes respectively. Further, AnnData saves whole matrices of calculations for the observations (obsm) or variables (varm) with the corresponding dimensions. Graph like structures which associate cells with cells or genes with genes are usually saved in obsp and varp. Any other unstructured data which does not fit any other slot is saved as unstructured data in uns. It is further possible to store more values of X in layers. Use cases for this are for example the storage of raw, unnormalized count data in a counts layer and the normalized data in the unnamed default layer.
AnnData is primarily designed for unimodal (for example just scRNA-Seq) data. However, extensions of AnnData such as MuData, which is covered later in this chapter, allow for the efficient storage and access of multimodal data.
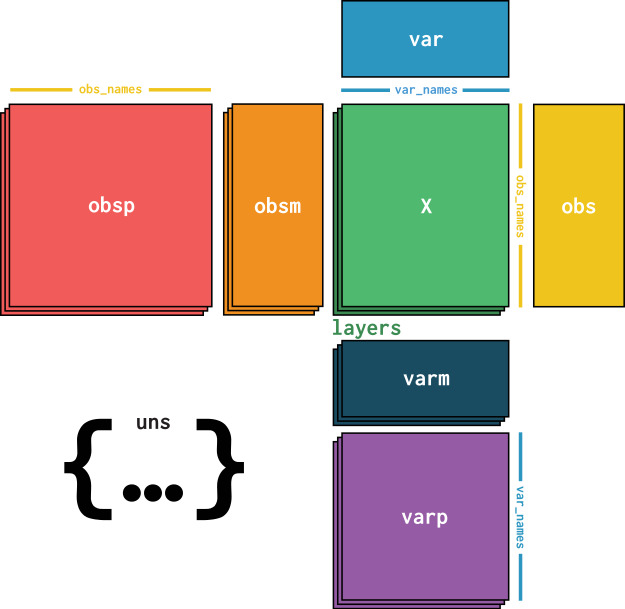
Fig. 4.1 AnnData overview. Image obtained from [Virshup et al., 2021].#
4.2.1. Installation#
AnnData is available on PyPI and Conda can be installed with using either:
pip install anndata
conda install -c conda-forge anndata
4.2.2. Initializing an AnnData object#
This section is inspired by AnnData’s “getting started” tutorial: https://anndata.readthedocs.io/en/latest/tutorials/notebooks/getting-started.html
Let us create a simple AnnData object with sparse count information which may for example represent gene expression counts. First, we import the required packages.
import anndata as ad
import numpy as np
import pandas as pd
from scipy.sparse import csr_matrix
As a next step we initialize an AnnData object with random poisson distributed data. It is an unwritten rule to name the primary AnnData object of the analysis adata.
counts = csr_matrix(
np.random.default_rng().poisson(1, size=(100, 2000)), dtype=np.float32
)
adata = ad.AnnData(counts)
adata
AnnData object with n_obs × n_vars = 100 × 2000
The obtained AnnData object has 100 observations and 2000 variables. This would correspond to 100 cells with 2000 genes. The initial data we passed are accessible as a sparse matrix using adata.X.
adata.X
<100x2000 sparse matrix of type '<class 'numpy.float32'>'
with 126413 stored elements in Compressed Sparse Row format>
Now, we provide the index to both the obs and var axes using .obs_names and .var_names respectively.
adata.obs_names = [f"Cell_{i:d}" for i in range(adata.n_obs)]
adata.var_names = [f"Gene_{i:d}" for i in range(adata.n_vars)]
print(adata.obs_names[:10])
Index(['Cell_0', 'Cell_1', 'Cell_2', 'Cell_3', 'Cell_4', 'Cell_5', 'Cell_6',
'Cell_7', 'Cell_8', 'Cell_9'],
dtype='object')
4.2.3. Adding aligned metadata#
4.2.3.1. Observational or Variable level#
The core of our AnnData object is now in place. As a next step we add metadata at both, the observational and variable levels. Remember, we store such annotations in the .obs and .var slots of the AnnData object for cell and gene annotations respectively.
ct = np.random.default_rng().choice(["B", "T", "Monocyte"], size=(adata.n_obs,))
adata.obs["cell_type"] = pd.Categorical(ct) # Categoricals are preferred for efficiency
adata.obs
| cell_type | |
|---|---|
| Cell_0 | B |
| Cell_1 | T |
| Cell_2 | B |
| Cell_3 | T |
| Cell_4 | Monocyte |
| ... | ... |
| Cell_95 | T |
| Cell_96 | T |
| Cell_97 | B |
| Cell_98 | T |
| Cell_99 | Monocyte |
100 rows × 1 columns
If we examine the representation of the AnnData object again now, we will notice that it got updated as well with the cell_type information in obs.
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'cell_type'
4.2.3.2. Subsetting using metadata#
We can also subset the AnnData object with the randomly generated cell types. The slicing and masking of the AnnData object behaves similarly to the access of data in Pandas DataFrames or R matrices. More details on this can be found below.
bdata = adata[adata.obs.cell_type == "B"]
bdata
View of AnnData object with n_obs × n_vars = 26 × 2000
obs: 'cell_type'
4.2.4. Observation/variable-level matrices#
We might also have metadata at either level that has many dimensions to it, such as a UMAP embedding of the data. For this type of metadata, AnnData has the .obsm/.varm attributes. We use keys to identify the different matrices we insert. The restriction of .obsm/.varm are that .obsm matrices must have a length equal to the number of observations as .n_obs and .varm matrices must have a length equal to .n_vars. They can each independently have different number of dimensions.
Let us start with a randomly generated matrix that we can interpret as a UMAP embedding of the data we would like to store, as well as some random gene-level metadata.
adata.obsm["X_umap"] = np.random.default_generator().normal(0, 1, size=(adata.n_obs, 2))
adata.varm["gene_stuff"] = np.random.default_generator().normal(
0, 1, size=(adata.n_vars, 5)
)
adata.obsm
AxisArrays with keys: X_umap
Again, the AnnData representation is updated.
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'cell_type'
obsm: 'X_umap'
varm: 'gene_stuff'
A few more notes about .obsm/.varm:
The “array-like” metadata can originate from a Pandas DataFrame, scipy sparse matrix, or numpy dense array.
When using scanpy, their values (columns) are not easily plotted, where instead items from
.obsare easily plotted on, e.g., UMAP plots.
4.2.5. Unstructured metadata#
As mentioned above, AnnData has .uns, which allows for any unstructured metadata. This can be anything, like a list or a dictionary with some general information that was useful in the analysis of our data. Try to only use this slot for data which cannot be efficiently stored in the other slots.
adata.uns["random"] = [1, 2, 3]
adata.uns
OverloadedDict, wrapping:
OrderedDict([('random', [1, 2, 3])])
With overloaded keys:
['neighbors'].
4.2.6. Layers#
Finally, we may have different forms of our original core data, perhaps one that is normalized and one that is not. These can be stored in different layers in AnnData. For example, let us log transform the original data and store it in a layer.
adata.layers["log_transformed"] = np.log1p(adata.X)
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'cell_type'
uns: 'random'
obsm: 'X_umap'
varm: 'gene_stuff'
layers: 'log_transformed'
Our original matrix X was not modified and is still accessible. We can verify this by comparing the original X to the new layer.
(adata.X != adata.layers["log_transformed"]).nnz == 0
False
4.2.7. Conversion to DataFrames#
It is possible to obtain a Pandas DataFrame from one of the layers.
adata.to_df(layer="log_transformed")
| Gene_0 | Gene_1 | Gene_2 | Gene_3 | Gene_4 | Gene_5 | Gene_6 | Gene_7 | Gene_8 | Gene_9 | ... | Gene_1990 | Gene_1991 | Gene_1992 | Gene_1993 | Gene_1994 | Gene_1995 | Gene_1996 | Gene_1997 | Gene_1998 | Gene_1999 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell_0 | 0.693147 | 0.000000 | 1.386294 | 1.386294 | 0.693147 | 0.000000 | 0.693147 | 1.098612 | 0.000000 | 1.098612 | ... | 0.693147 | 0.693147 | 0.000000 | 0.693147 | 1.098612 | 0.000000 | 0.000000 | 1.098612 | 0.693147 | 0.000000 |
| Cell_1 | 1.098612 | 1.098612 | 0.000000 | 0.000000 | 0.000000 | 0.693147 | 0.000000 | 0.693147 | 1.386294 | 0.000000 | ... | 1.098612 | 1.386294 | 1.098612 | 0.000000 | 0.693147 | 0.693147 | 1.386294 | 0.000000 | 0.000000 | 0.693147 |
| Cell_2 | 0.693147 | 0.693147 | 1.098612 | 0.693147 | 1.386294 | 1.386294 | 0.693147 | 0.693147 | 0.000000 | 1.098612 | ... | 0.000000 | 0.693147 | 0.693147 | 0.000000 | 1.386294 | 0.693147 | 0.000000 | 0.000000 | 0.693147 | 0.693147 |
| Cell_3 | 0.000000 | 0.000000 | 0.000000 | 0.693147 | 0.693147 | 0.000000 | 1.386294 | 1.098612 | 0.693147 | 0.693147 | ... | 0.693147 | 0.693147 | 0.000000 | 0.693147 | 0.693147 | 0.693147 | 0.693147 | 0.000000 | 1.386294 | 0.693147 |
| Cell_4 | 0.000000 | 0.693147 | 0.000000 | 1.609438 | 1.098612 | 0.693147 | 0.000000 | 0.000000 | 0.693147 | 0.000000 | ... | 1.098612 | 1.098612 | 0.000000 | 1.386294 | 1.791759 | 1.098612 | 1.609438 | 0.693147 | 0.000000 | 0.693147 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Cell_95 | 0.693147 | 1.098612 | 0.693147 | 1.386294 | 1.098612 | 0.693147 | 0.000000 | 0.000000 | 0.000000 | 0.693147 | ... | 0.000000 | 0.693147 | 0.000000 | 0.000000 | 0.000000 | 0.693147 | 0.000000 | 0.693147 | 1.386294 | 0.693147 |
| Cell_96 | 0.693147 | 0.000000 | 0.000000 | 1.098612 | 0.000000 | 0.693147 | 0.693147 | 1.098612 | 0.000000 | 0.000000 | ... | 1.386294 | 0.693147 | 1.098612 | 0.693147 | 0.000000 | 0.693147 | 1.098612 | 1.386294 | 0.693147 | 0.000000 |
| Cell_97 | 0.693147 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.098612 | 0.000000 | 0.000000 | 0.693147 | ... | 0.000000 | 0.693147 | 1.386294 | 0.000000 | 0.000000 | 0.000000 | 0.693147 | 0.693147 | 1.609438 | 0.693147 |
| Cell_98 | 0.693147 | 0.693147 | 0.000000 | 1.098612 | 0.000000 | 0.693147 | 0.693147 | 1.098612 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.693147 | 0.693147 | 0.000000 | 0.693147 | 1.386294 | 0.693147 | 1.098612 | 1.386294 |
| Cell_99 | 1.098612 | 1.791759 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.098612 | 1.609438 | 0.693147 | 0.693147 | ... | 0.693147 | 0.000000 | 0.000000 | 0.693147 | 0.693147 | 0.693147 | 0.693147 | 0.000000 | 0.000000 | 1.386294 |
100 rows × 2000 columns
4.2.8. Reading and writing of AnnData objects#
AnnData objects can be saved on disk to hierarchical array stores like HDF5 or Zarr to enable similar structures in disk and on memory. AnnData comes with its own persistent HDF5-based file format: h5ad. If string columns with small number of categories are not yet categoricals, AnnData will auto-transform them to categoricals. We will now save our AnnData object in h5ad format.
adata.write("my_results.h5ad", compression="gzip")
!h5ls 'my_results.h5ad'
X Group
layers Group
obs Group
obsm Group
uns Group
var Group
varm Group
… and read it back in.
adata_new = ad.read_h5ad("my_results.h5ad")
adata_new
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'cell_type'
uns: 'random'
obsm: 'X_umap'
varm: 'gene_stuff'
layers: 'log_transformed'
4.2.9. Efficient data access#
4.2.9.1. View and copies#
For the fun of it, let us look at another metadata use case. Imagine that the observations come from instruments characterizing 10 readouts in a multi-year study with samples taken from different subjects at different sites. We would typically get that information in some format and then store it in a DataFrame:
obs_meta = pd.DataFrame(
{
"time_yr": np.random.default_rng().choice([0, 2, 4, 8], adata.n_obs),
"subject_id": np.random.default_rng().choice(
["subject 1", "subject 2", "subject 4", "subject 8"], adata.n_obs
),
"instrument_type": np.random.default_rng().choice(
["type a", "type b"], adata.n_obs
),
"site": np.random.default_rng().choice(["site x", "site y"], adata.n_obs),
},
index=adata.obs.index, # these are the same IDs of observations as above!
)
This is how we join the readout data with the metadata. Of course, the first argument of the following call for X could also just be a DataFrame. This will result in a single data container which tracks everything.
adata = ad.AnnData(adata.X, obs=obs_meta, var=adata.var)
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'time_yr', 'subject_id', 'instrument_type', 'site'
Subsetting the joint data matrix can be important to focus on subsets of variables or observations, or to define train-test splits for a machine learning model.
Note Similar to numpy arrays, AnnData objects can either hold actual data or reference another AnnData object. In the later case, they are referred to as “view”. Subsetting AnnData objects always returns views, which has two advantages:
no new memory is allocated
it is possible to modify the underlying AnnData object.
You can get an actual AnnData object from a view by calling .copy() on the view. Usually, this is not necessary, as any modification of elements of a view (calling .[] on an attribute of the view) internally calls .copy() and makes the view an AnnData object that holds actual data. See the example below.
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'time_yr', 'subject_id', 'instrument_type', 'site'
Note Indexing into AnnData will assume that integer arguments to [] behave like .iloc in pandas, whereas string arguments behave like .loc. AnnData always assumes string indices.
adata_view = adata[:5, ["Gene_1", "Gene_3"]]
adata_view
View of AnnData object with n_obs × n_vars = 5 × 2
obs: 'time_yr', 'subject_id', 'instrument_type', 'site'
This is a view! This can be verified by examining the AnnData object again.
adata
AnnData object with n_obs × n_vars = 100 × 2000
obs: 'time_yr', 'subject_id', 'instrument_type', 'site'
The dimensions of the AnnData object have not changed it still contains the same data. If we want an AnnData that holds the data in memory we have to call .copy().
adata_subset = adata[:5, ["Gene_1", "Gene_3"]].copy()
For a view, we can also set the first 3 elements of a column.
print(adata[:3, "Gene_1"].X.toarray().tolist())
adata[:3, "Gene_1"].X = [0, 0, 0]
print(adata[:3, "Gene_1"].X.toarray().tolist())
[[0.0], [2.0], [1.0]]
[[0.0], [0.0], [0.0]]
If you try to access parts of a view of an AnnData, the content will be auto-copied and a data-storing object will be generated.
adata_subset = adata[:3, ["Gene_1", "Gene_2"]]
adata_subset
View of AnnData object with n_obs × n_vars = 3 × 2
obs: 'time_yr', 'subject_id', 'instrument_type', 'site'
adata_subset.obs["foo"] = range(3)
Trying to set attribute `.obs` of view, copying.
Now adata_subset stores the actual data and is no longer just a reference to adata.
adata_subset
AnnData object with n_obs × n_vars = 3 × 2
obs: 'time_yr', 'subject_id', 'instrument_type', 'site', 'foo'
Evidently, you can use all of pandas to slice with sequences or boolean indices.
adata[adata.obs.time_yr.isin([2, 4])].obs.head()
| time_yr | subject_id | instrument_type | site | |
|---|---|---|---|---|
| Cell_0 | 4 | subject 8 | type b | site x |
| Cell_2 | 2 | subject 2 | type b | site y |
| Cell_3 | 4 | subject 8 | type a | site y |
| Cell_5 | 2 | subject 2 | type a | site y |
| Cell_10 | 2 | subject 8 | type b | site x |
4.2.9.2. Partial reading of large data#
If a single h5ad file is very large, you can partially read it into memory by using backed mode or with the currently experimental read_elem API.
adata = ad.read("my_results.h5ad", backed="r")
adata.isbacked
True
If you do this, you will need to remember that the AnnData object has an open connection to the file used for reading.
adata.filename
PosixPath('my_results.h5ad')
As we are using it in read-only mode, we cannot damage anything. To proceed with this tutorial, we still need to explicitly close it.
adata.file.close()
4.3. Unimodal data analysis with scanpy#
Now that we understood the fundamental data structure of unimodal single-cell analysis, the question remains how we can actually analyze the stored data. In the scverse ecosystem several tools for the analysis of specific omics data exist. For example, scanpy[Wolf et al., 2018] provides tooling for general RNA-Seq focused analysis, squidpy[Palla et al., 2022] focuses on spatial transcriptomics and scirpy[Sturm et al., 2020] provides tooling for the analysis of T-cell receptor (TCR) and B-cell receptor (BCR) data. Even though many scverse extensions for various data modalities exist, they usually use some of scanpy’s preprocessing and visualization capabilities to some extent.
More specifically, scanpy is a Python package which builds on top of AnnData to facilitate the analysis of single-cell gene expression data. Several methods for preprocessing, embedding, visualization, clustering, differential gene expression testing, pseudotime and trajectory inference, and simulation of gene regulatory networks are accessible through scanpy. The efficient implementation based on the Python data science and machine learning libraries allows scanpy to scale to millions of cells.
Generally, best-practice single-cell data analysis is an interactive process. Many of the decisions and analysis steps depend on the results of previous steps and potentially input of experimental partners. Pipelines such as scflow[Khozoie et al., 2021], which fully automate some downstream analysis steps, started to appear only recently. These pipelines have to make assumptions and simplifications which may not result in the most robust analysis.
scanpy is therefore designed to be used for interactive analyses with for example Jupyter Notebooks[Jupyter, 2022].
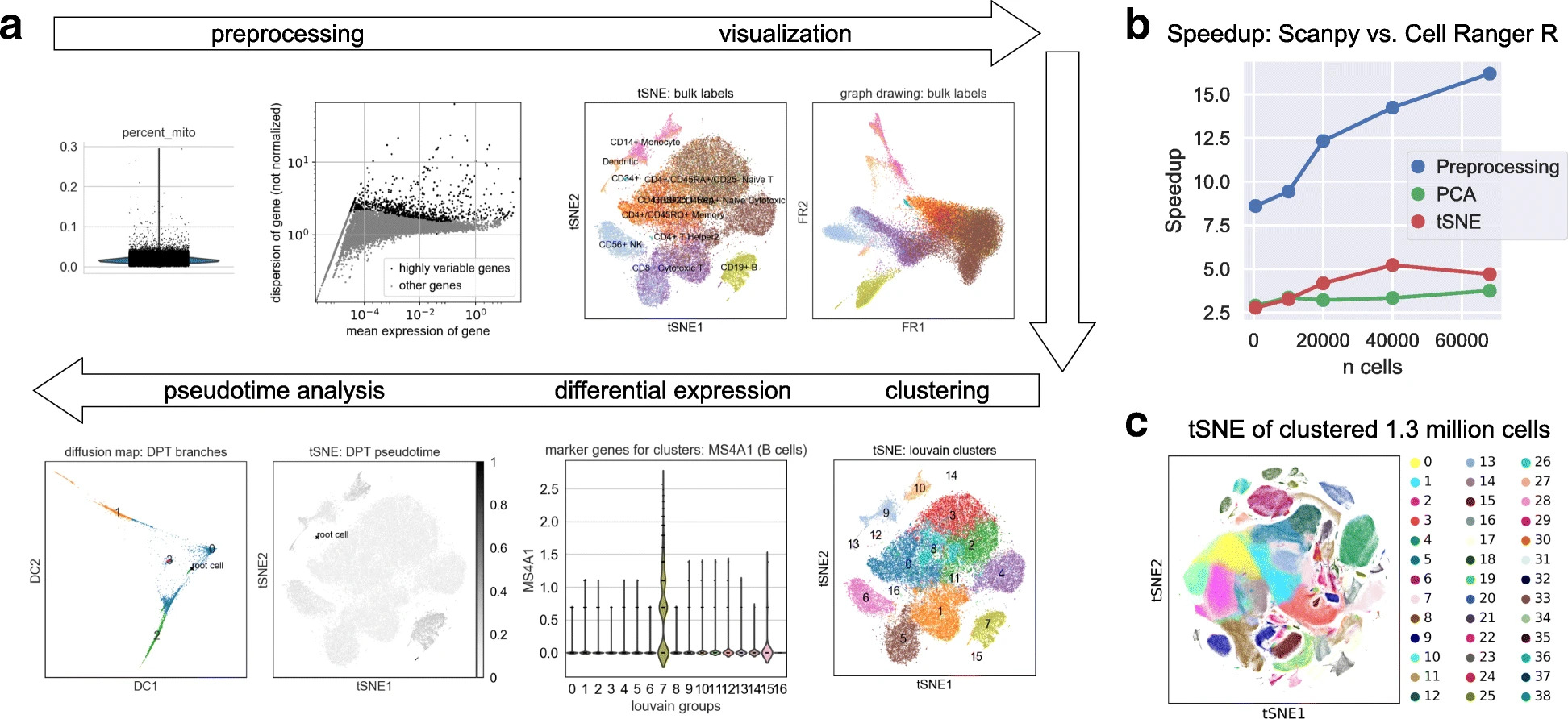
Fig. 4.2 scanpy overview. Image obtained from [Wolf et al., 2018].#
4.3.1. Installation#
scanpy is available on PyPI and Conda can be installed with using either:
pip install scanpy
conda install -c conda-forge scanpy
4.3.2. Scanpy API design#
The scanpy framework is designed in a way that functions belonging to the same step are grouped in corresponding modules. For example, all preprocessing functions are available in the scanpy.preprocessing module, all transformation of a data matrix which are not preprocessing are available in scanpy.tools and all visualizations are available in scanpy.plot. These modules are commonly accessed after having imported scanpy like import scanpy as sc with the corresponding abbreviations sc.pp for preprocessing, sc.tl for tools and sc.pl for plots. All modules which read or write data are directly accessed. Further, a module for various datasets is available as sc.datasets.
All functions with their corresponding parameters and potentially example plots are documented in the scanpy API documentation[scverse scanpy, 2022].
Note that this tutorial only covers a very small subset of scanpy’s features and options. Readers are strongly encouraged to examine scanpy’s documentation for more details.
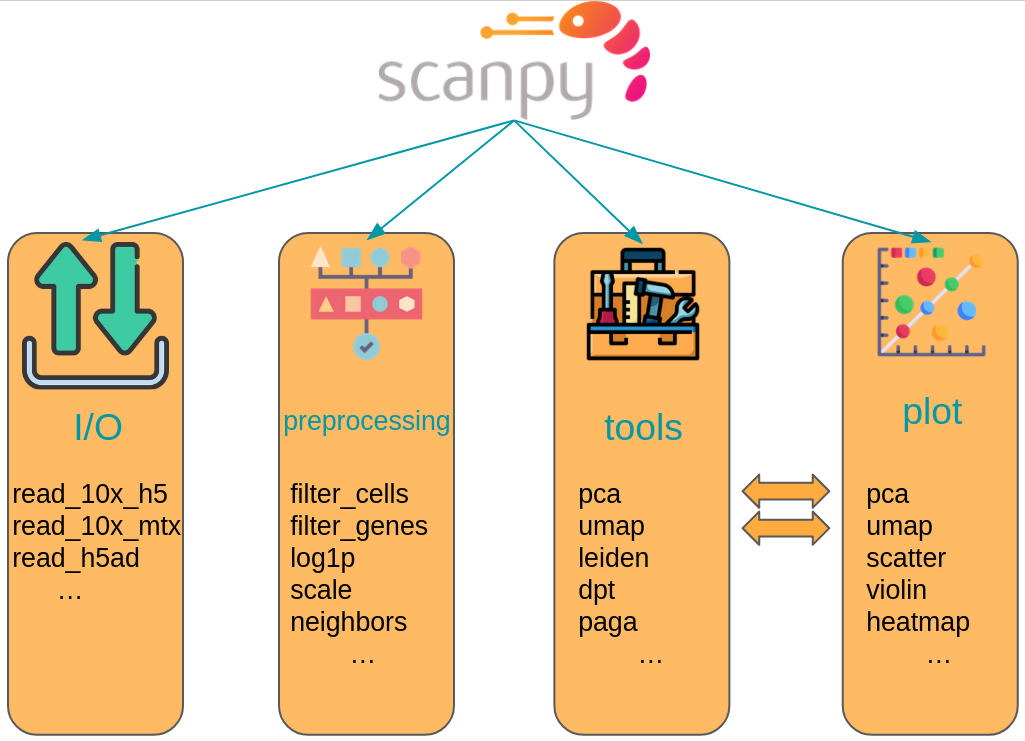
Fig. 4.3 scanpy API overview. The API is divided into datasets, preprocessing (pp), tools (tl) and corresponding plotting (pl) functions.#
4.3.3. Scanpy example#
In the following cells we will shortly demonstrate the workflow of an analysis with scanpy. We explicitly do not conduct a full analysis because the specific analysis steps are covered in the corresponding chapters.
The dataset of choice is a dataset of 2700 peripheral blood mononuclear cells of a healthy donor which were sequenced on the Illumina NextSeq 500.
As a first step we import scanpy and define defaults for our following quick scanpy demo. We use scanpy’s setting object to set the Matplotlib plotting defaults for all of scanpy’s plots and finally print scanpy’s header. This header contains the versions of all relevant Python packages in the current environment including Scanpy and AnnData. This output is especially useful when reporting bugs to the scverse team and for reproducibility reasons.
import scanpy as sc
sc.settings.set_figure_params(dpi=80, facecolor="white")
sc.logging.print_header()
scanpy==1.9.1 anndata==0.7.8 umap==0.5.2 numpy==1.20.3 scipy==1.7.2 pandas==1.4.3 scikit-learn==1.0.1 statsmodels==0.13.1 python-igraph==0.9.10 pynndescent==0.5.5
Next, we get the 3k PBMC dataset with scanpy as an AnnData object. The dataset is automatically downloaded and saved to a path which can be also specified with scanpy’s settings object. By default it is the folder data.
adata = sc.datasets.pbmc3k()
adata
100%|██████████| 5.58M/5.58M [00:01<00:00, 3.25MB/s]
AnnData object with n_obs × n_vars = 2700 × 32738
var: 'gene_ids'
The returned AnnData object has 2700 cells with 32738 genes. The var slot further contains the gene IDs.
adata.var
| gene_ids | |
|---|---|
| index | |
| MIR1302-10 | ENSG00000243485 |
| FAM138A | ENSG00000237613 |
| OR4F5 | ENSG00000186092 |
| RP11-34P13.7 | ENSG00000238009 |
| RP11-34P13.8 | ENSG00000239945 |
| ... | ... |
| AC145205.1 | ENSG00000215635 |
| BAGE5 | ENSG00000268590 |
| CU459201.1 | ENSG00000251180 |
| AC002321.2 | ENSG00000215616 |
| AC002321.1 | ENSG00000215611 |
32738 rows × 1 columns
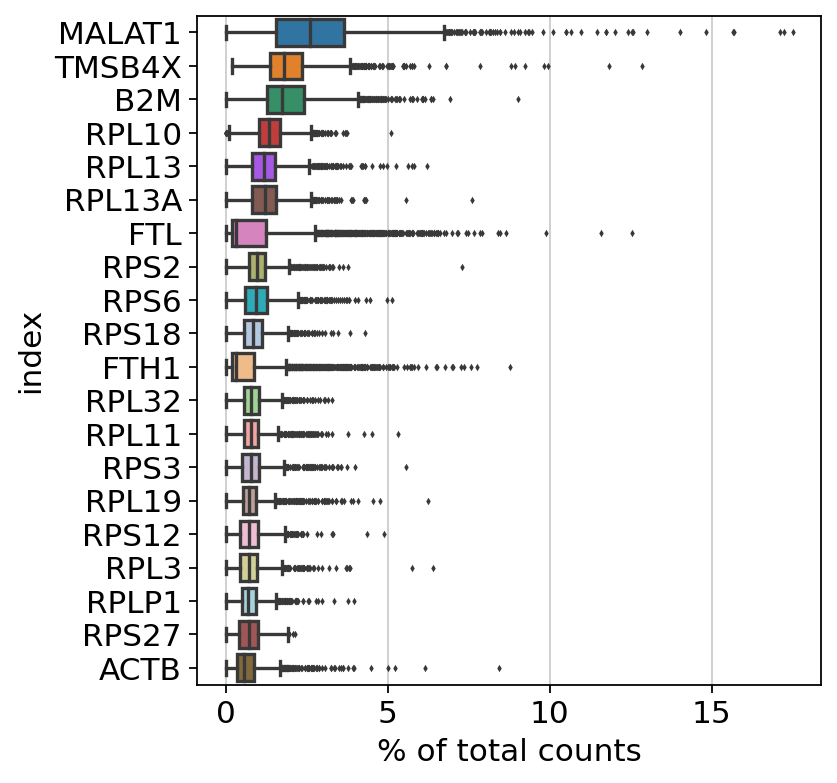
As mentioned above, all of scanpy’s analysis functions are accessible via sc.[pp, tl, pl]. As a first step to get an overview over our data, we use scanpy to show those genes that yield the highest fraction of counts in each single cell, across all cells. We simply call the sc.pl.highest_expr_genes function, pass the AnnData object which is in pretty much all cases the first parameter of any scanpy function, and specify that we want the top 20 expressed genes to be shown.
Apparently, MALAT1 is the most expressed gene which is frequently detected in poly-A captured scRNA-Seq data, independent of protocol. This gene has been shown to have an inverse correlation with cell health. Especially dead/dying cells have a higher expression of MALAT1.
We now filter cells with less than 200 detected genes and genes which were found in less than 3 cells for a rough quality threshold with scanpy’s preprocessing module.
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
A common step in single-cell RNA-Seq analysis is dimensionality reduction with for example principal component analysis (PCA) to unveil the main axes of variation. This also denoises the data. scanpy offers PCA as a preprocessing or tools function. These are equivalent. Here, we use the version in tools for no particular reason.
sc.tl.pca(adata, svd_solver="arpack")
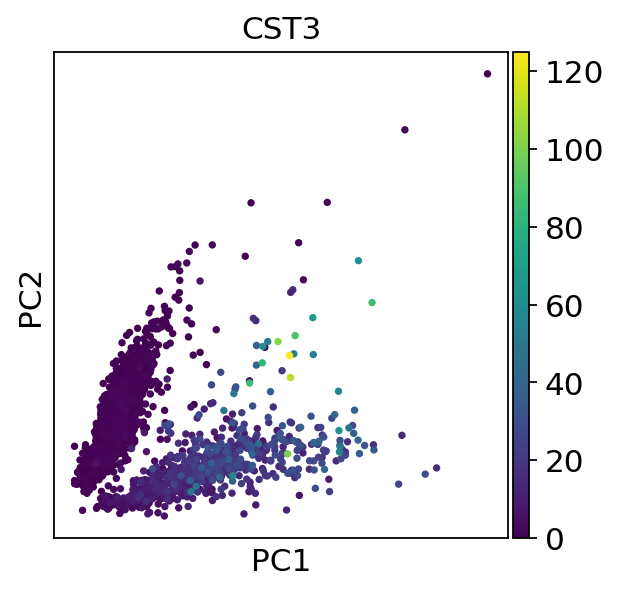
The corresponding plotting function allows us to pass genes to the color argument. The corresponding values are automatically extracted from the AnnData object.
A fundamental step for any advanced embedding and downstream calculations is the calculating of the neighborhood graph using the PCA representation of the data matrix. It is automatically used for other tools that require it such as the calculation of a UMAP.
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
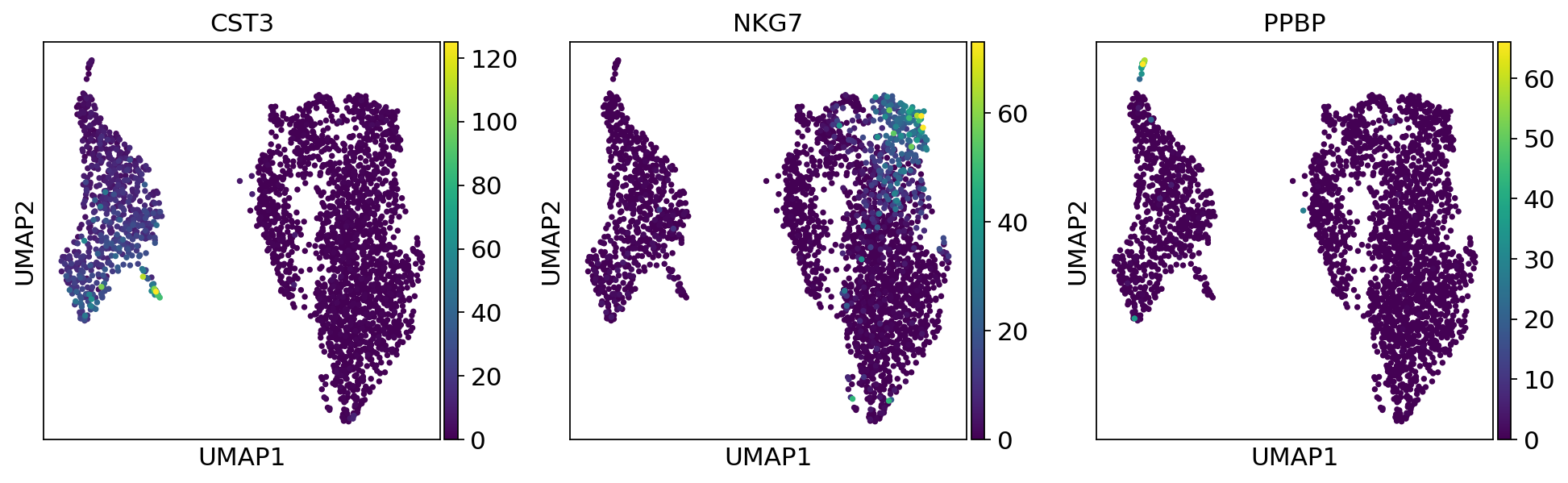
We now use the calculating neighborhood graph to embed the cells with a UMAP, one of many advanced dimension reduction algorithms implemented in scanpy.
sc.tl.umap(adata)
scanpy’s documentation also provides tutorials which we recommend to all readers who need a refresher of scanpy or are new to scanpy. Video tutorials are available on the scverse youtube channel.
4.4. Advanced: Using MuData to store multimodal data#
AnnData is primarily designed for storing and manipulating unimodal data. However, multimodal assays such as CITE-Seq generate multimodal data by simultaneously measuring RNA and surface proteins. This data requires more advanced ways of storing, which is where MuData comes into play. MuData builds on top of AnnData to store and manipulate multimodal data. muon[Bredikhin et al., 2022], a “scanpy equivalent” and core package of scverse, can then be used to analyze the multimodal omics data. The following section is based on the MuData Quickstart tutorial[scverse mudata, 2022] and the Multimodal data objects tutorial[scverse mudata, 2022].
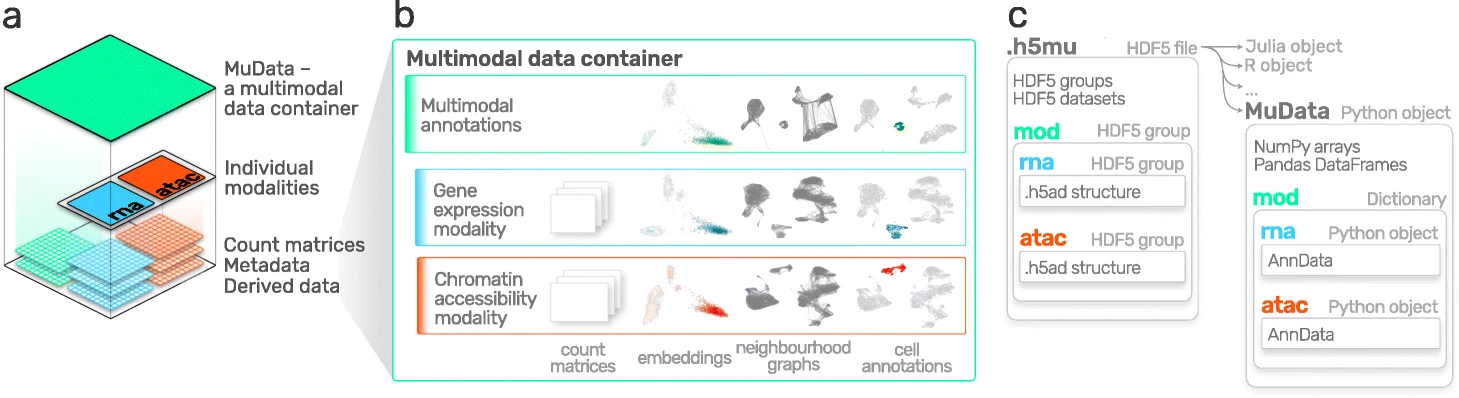
Fig. 4.4 MuData overview. Image obtained from [Bredikhin et al., 2022].#
4.4.1. Installation#
MuData is available on PyPI and Conda can be installed with using either:
pip install mudata
conda install -c conda-forge mudata
The main idea behind MuData is that the MuData object contains references to the single AnnData objects of the unimodal data, but the MuData object itself also stores multimodal annotations. It is therefore possible to directly access the AnnData objects to perform unimodal data transformations which store their results in the corresponding AnnData annotations, but also to aggregate the modalities for joint calculations whose results can be stored in the global MuData object. Technically, this is realized by MuData objects comprising a dictionary with AnnData objects, one per modality, in their .mod (=modality) attribute. Just as AnnData objects themselves, they also contain attributes like .obs with annotation of observations (samples or cells), .obsm with their multidimensional annotations such as embeddings.
4.4.2. Initializing a MuData object#
We will start by importing MuData from the mudata package.
import mudata as md
To create an example MuData object we require simulated data.
n, d, k = 1000, 100, 10
z = np.random.default_generator().normal(
loc=np.arange(k), scale=np.arange(k) * 2, size=(n, k)
)
w = np.random.default_generator().normal(size=(d, k))
y = np.dot(z, w.T)
y.shape
(1000, 100)
To create a MuData object we require the unimodal AnnData objects first. Therefore, we create two AnnData objects with data for the same observations, but for different variables.
adata = ad.AnnData(y)
adata.obs_names = [f"obs_{i+1}" for i in range(n)]
adata.var_names = [f"var_{j+1}" for j in range(d)]
adata
AnnData object with n_obs × n_vars = 1000 × 100
d2 = 50
w2 = np.random.default_generator().normal(size=(d2, k))
y2 = np.dot(z, w2.T)
adata2 = ad.AnnData(y2)
adata2.obs_names = [f"obs_{i+1}" for i in range(n)]
adata2.var_names = [f"var2_{j+1}" for j in range(d2)]
adata2
AnnData object with n_obs × n_vars = 1000 × 50
These two AnnData objects (two “modalities”) can then be wrapped into a single MuData object. Here, we name modality one A and modality two B.
mdata = md.MuData({"A": adata, "B": adata2})
mdata
MuData object with n_obs × n_vars = 1000 × 150
2 modalities
A: 1000 x 100
B: 1000 x 50Observations and variables of the MuData object are global, which means that observations with the identical name (.obs_names) in different modalities are considered to be the same observation. This also means variable names (.var_names) should be unique. This is reflected in the object description above: mdata has 1000 observations and 150 = 100+50 variables.
4.4.3. MuData attributes#
MuData objects consist of annotations as earlier described for AnnData objects like .obs or .var, but extend this behavior with .mod which serves as an accessor to the individual modalities.
Modalities are stored in a collection accessible via the .mod attribute of the MuData object with names of modalities as keys and AnnData objects as values.
list(mdata.mod.keys())
['A', 'B']
Individual modalities can be accessed with their names via the .mod attribute or via the MuData object itself as a shorthand.
print(mdata.mod["A"])
print(mdata["A"])
AnnData object with n_obs × n_vars = 1000 × 100
AnnData object with n_obs × n_vars = 1000 × 100
Samples (cells) annotation is accessible via the .obs attribute and by default includes copies of columns from .obs data frames of individual modalities. The same goes for .var, which contains annotation of variables (features). Observations columns copied from individual modalities contain modality name as their prefix, e.g. rna:n_genes. This is also true for variables columns. However if there are columns with identical names in .var of multiple modalities — e.g. n_cells, — these columns are merged across modalities and no prefix is added. When those slots are changed in AnnData objects of modalities, e.g. new columns are added or samples (cells) are filtered out, the changes have to be fetched with the .update() method (see below).
Multidimensional annotations of samples (cells) are accessible in the .obsm attribute. For instance, that can be UMAP coordinates that were learnt jointly on all modalities.
The MuData object’s shape is represented by two numbers calculated as a sum of the shapes of individual modalities — one for the number of observations and one for the number of variables.
print(mdata.shape)
print(mdata.n_obs)
print(mdata.n_vars)
(1000, 150)
1000
150
By default, variables are always counted as belonging uniquely to a single modality while observations with the same name are counted as the same observation, which has variables across multiple modalities measured for.
[adata.shape for adata in mdata.mod.values()]
[(1000, 100), (1000, 50)]
If the shape of a modality is changed, MuData.update() has to be run to bring the respective updates to the MuData object.
adata2.var_names = ["var_ad2_" + e.split("_")[1] for e in adata2.var_names]
print("Outdated variables names: ...,", ", ".join(mdata.var_names[-3:]))
mdata.update()
print("Updated variables names: ...,", ", ".join(mdata.var_names[-3:]))
Outdated variables names: ..., var2_48, var2_49, var2_50
Updated variables names: ..., var_ad2_48, var_ad2_49, var_ad2_50
Importantly, individual modalities are stored as references to the original objects. Hence, if the original AnnData is changed the change will also be reflected in the MuData object.
# Add some unstructured data to the original object
adata.uns["misc"] = {"adata": True}
# Access modality A via the .mod attribute
mdata.mod["A"].uns["misc"]
{'adata': True}
4.4.4. Variable mappings#
Upon construction of a MuData object, a global binary mapping between observations and individual modalities is created as well as between variables and modalities. Since all the observations are the same across modalities in mdata, all the values in the observations mappings are set to True.
np.sum(mdata.obsm["A"]) == np.sum(mdata.obsm["B"]) == n
True
For variables however, those are 150-long vectors. The A modality has 100 True values followed by 50 False values.
mdata.varm["A"]
array([ True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, True, True, True, True, True, True, True, True,
True, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False])
4.4.5. MuData views#
Analogous to the behavior of AnnData objects, slicing MuData objects returns views of the original data.
view = mdata[:100, :1000]
print(view.is_view)
print(view["A"].is_view)
True
True
Subsetting MuData objects is special since it slices them across modalities. I.e. the slicing operation for a set of obs_names and/or var_names will be performed for each modality and not only for the global multimodal annotation. This behavior makes workflows memory-efficient, which is especially important when working with large datasets. If the object is to be modified however, a copy of it should be created, which is not a view anymore and has no dependence on the original object.
mdata_sub = view.copy()
mdata_sub.is_view
False
4.4.6. Common observations#
While a MuData object is comprised of the same observations for both modalities, it is not always the case in the real world where some data might be missing. By design, MuData accounts for these scenarios since there’s no guarantee observations are the same — or even intersecting — for a MuData instance. It’s worth noting that other tools might provide convenience functions for some common scenarios of dealing with missing data, such as intersect_obs() implemented in muon.
4.4.7. Reading and Writing of MuData objects#
Similarly to AnnData objects, MuData objects were designed to be serialized into HDF5 based .h5mu files. All modalities are stored under their respective names in the /mod HDF5 group of the .h5mu file. Each individual modality, e.g. /mod/A, is stored in the same way as it would be stored in the .h5ad file. MuData objects can be read and written as follows:
mdata.write("my_mudata.h5mu")
mdata_r = md.read("my_mudata.h5mu", backed=True)
mdata_r
MuData object with n_obs × n_vars = 1000 × 150 backed at 'my_mudata.h5mu'
2 modalities
A: 1000 x 100
uns: 'misc'
B: 1000 x 50Individual modalities are backed as well inside the .h5mu file.
mdata_r["A"].isbacked
True
If the original object is backed, the filename has to be provided to the .copy() call, and the resulting object will be backed at a new location.
mdata_sub = mdata_r.copy("mdata_sub.h5mu")
print(mdata_sub.is_view)
print(mdata_sub.isbacked)
False
True
4.4.8. Multimodal methods#
When the MuData object is prepared, it is up to multimodal methods to be used to make sense of the data. The most simple and naïve approach is to concatenate matrices from multiple modalities to perform for example dimensionality reduction.
x = np.hstack([mdata.mod["A"].X, mdata.mod["B"].X])
x.shape
(1000, 150)
We can write a simple function to run principal component analysis on such a concatenated matrix. MuData object provides a place to store multimodal embeddings: MuData.obsm. It is similar to how the embeddings generated on individual modalities are stored, only this time it is saved inside the MuData object rather than in AnnData.obsm.
To calculate for example a principal component analysis (PCA) for the joint values of the modalities, we horizontally stack the values stored in the individual modalities and then perform the PCA on the stacked matrix. This is possible because the number of observations matches across modalities (remember, the number of features per modality does not have to match).
def simple_pca(mdata):
from sklearn import decomposition
x = np.hstack([m.X for m in mdata.mod.values()])
pca = decomposition.PCA(n_components=2)
components = pca.fit_transform(x)
# By default, methods operate in-place and embeddings are stored in the .obsm slot
mdata.obsm["X_pca"] = components
simple_pca(mdata)
print(mdata)
MuData object with n_obs × n_vars = 1000 × 150
obsm: 'X_pca'
2 modalities
A: 1000 x 100
uns: 'misc'
B: 1000 x 50
Our calculated principal components are now stored in the MuData object itself and accessible for further multimodal transformations.
In reality, however, having different modalities often means that the features between them come from different generative processes and are not comparable. This is where special multimodal integration methods come into play. For omics technologies, these methods are frequently addressed as multi-omics integration methods. In the following section we will introduce muon which provides many tools to preprocess unimodal data beyond RNA-Seq and multi-omics integration methods.
4.5. Advanced: Multimodal data analysis with muon#
Although scanpy provides generally applicable tools such as PCA, UMAP and various visualizations, it is primarily designed for the analysis of RNA-Seq data. muon fills this gap by providing preprocessing functions for other omics such as chromatin accessibility (ATAC) or protein (CITE) data. As mentioned above, muon further provides algorithms to run multi-omics algorithms which infer knowledge from the joint modalities. For example, users may run a PCA on a single modality, but muon further provides multi-omics factor analysis algorithms which take several modalities as input.
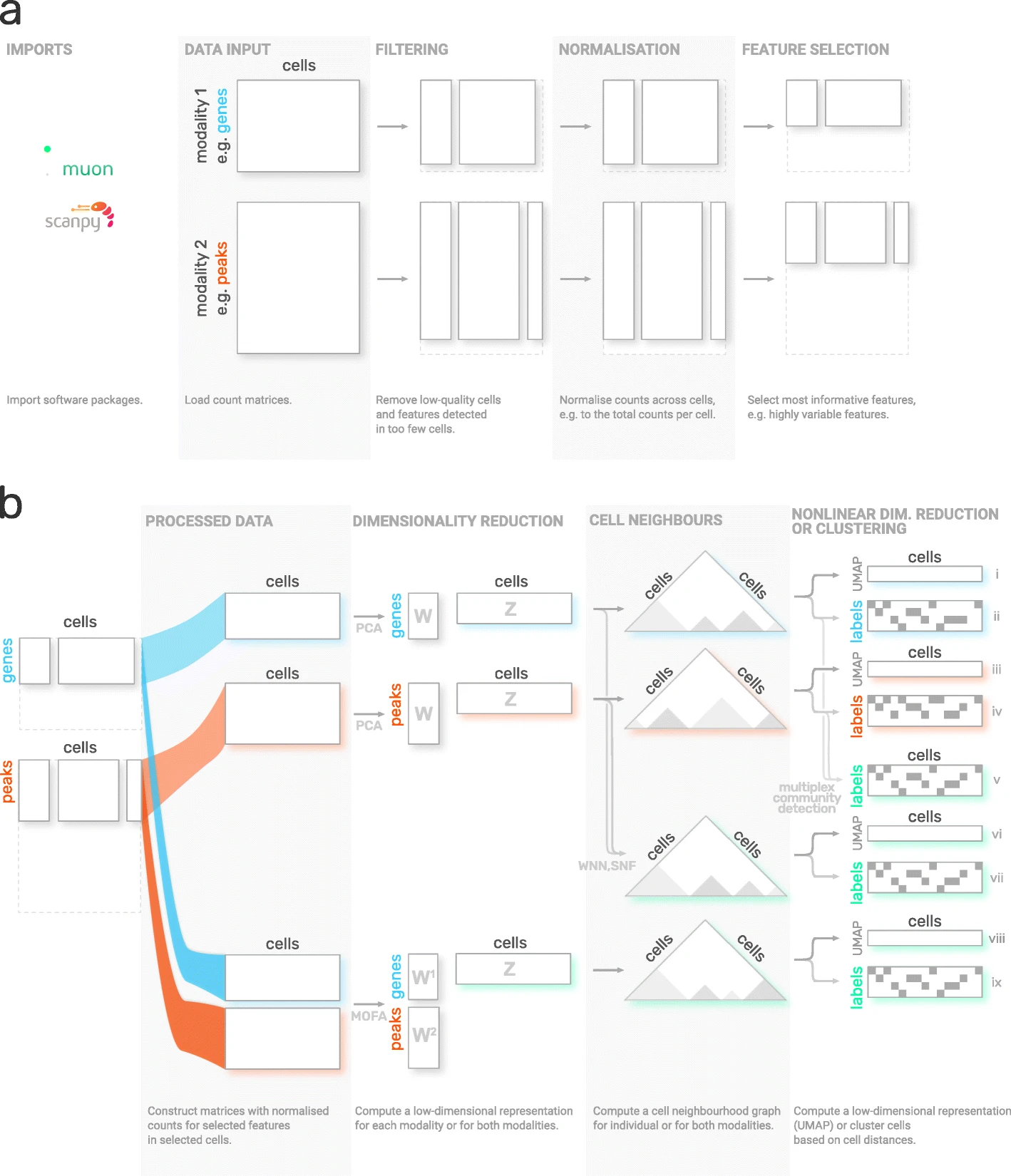
Fig. 4.5 muon overview. Image obtained from [Bredikhin et al., 2022].#
4.5.1. Installation#
muon is available on PyPI can be installed using:
pip install muon
4.5.2. API overview#
For the sake of introducing muon we will examine the ATAC data of a multimodal dataset. Analogously to the scanpy chapter, this chapter solely serves as a quick demo and overview over muon and does not analyze a dataset completely, let alone providing best-practice multi-omics analysis. Please read the corresponding chapters to learn how to properly conduct such analyses.
muon separates its modules in two ways. First, analogously to scanpy, general and multimodal functions are grouped in preprocessing (muon.pp), tools (muon.tl) and plots (muon.pl). Second, unimodal tools are available from the corresponding of muon, which are itself again separated into preprocessing, tools and plots. For example, all ATAC preprocessing functions are grouped into muon.atac.pp. This also applies to CITE-Seq preprocessing functions (muon.prot.pp).
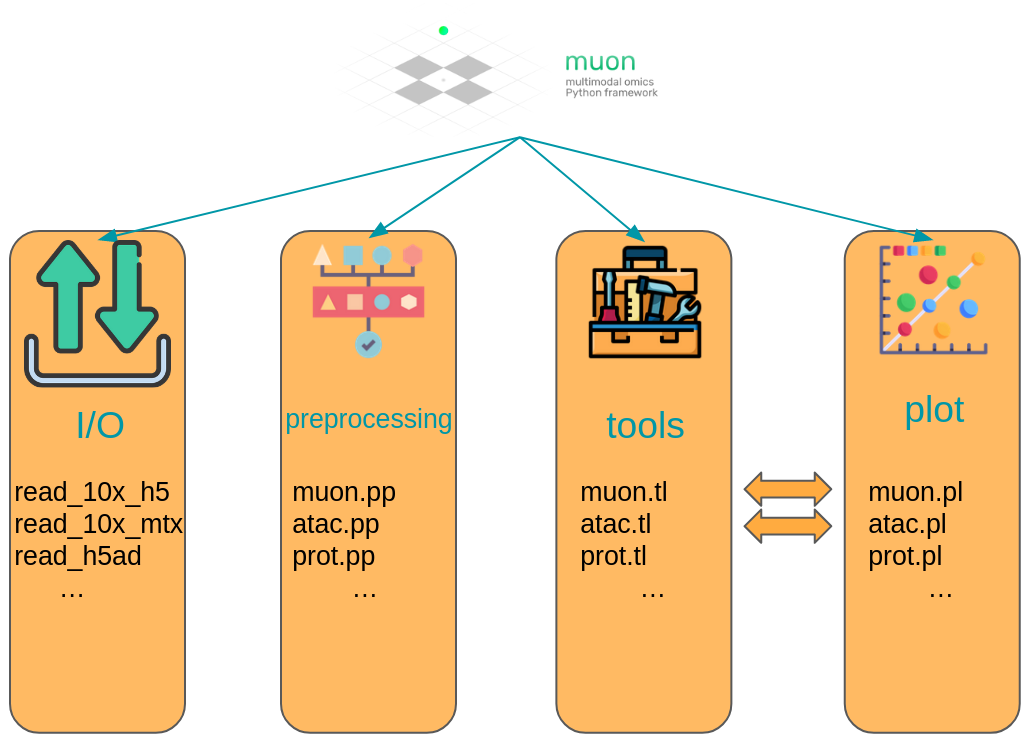
Fig. 4.6 muon API overview. Modality specific functions are provided in correspondingly named modules. Functions for the joint analysis of modalities are available via the muon module directly.#
4.5.3. muon API demo#
The dataset for our demo is a publicly available 10x Genomics Multiome dataset for human PBMCs. Let us import muon and get the dataset with mudatasets.
import mudatasets as mds
import muon as mu
mdata, _ = mds.load("pbmc10k_multiome", with_info=True, full=True)
mdata.var_names_make_unique()
■ File filtered_feature_bc_matrix.h5 from pbmc10k_multiome has been found at /home/zeth/mudatasets/pbmc10k_multiome/filtered_feature_bc_matrix.h5
■ Checksum is validated (md5) for filtered_feature_bc_matrix.h5
■ File atac_fragments.tsv.gz from pbmc10k_multiome has been found at /home/zeth/mudatasets/pbmc10k_multiome/atac_fragments.tsv.gz
■ Checksum is validated (md5) for atac_fragments.tsv.gz
■ File atac_fragments.tsv.gz.tbi from pbmc10k_multiome has been found at /home/zeth/mudatasets/pbmc10k_multiome/atac_fragments.tsv.gz.tbi
■ Checksum is validated (md5) for atac_fragments.tsv.gz.tbi
■ File atac_peaks.bed from pbmc10k_multiome has been found at /home/zeth/mudatasets/pbmc10k_multiome/atac_peaks.bed
■ Checksum is validated (md5) for atac_peaks.bed
■ File atac_peak_annotation.tsv from pbmc10k_multiome has been found at /home/zeth/mudatasets/pbmc10k_multiome/atac_peak_annotation.tsv
■ Checksum is validated (md5) for atac_peak_annotation.tsv
■ Loading filtered_feature_bc_matrix.h5...
/home/zeth/miniconda3/envs/bp2/lib/python3.9/site-packages/mudatasets/core.py:203: UserWarning: Dataset is in the 10X .h5 format and can't be loaded as backed.
warn("Dataset is in the 10X .h5 format and can't be loaded as backed.")
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Added `interval` annotation for features from /home/zeth/mudatasets/pbmc10k_multiome/filtered_feature_bc_matrix.h5
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Added peak annotation from /home/zeth/mudatasets/pbmc10k_multiome/atac_peak_annotation.tsv to .uns['atac']['peak_annotation']
Added gene names to peak annotation in .uns['atac']['peak_annotation']
Located fragments file: /home/zeth/mudatasets/pbmc10k_multiome/atac_fragments.tsv.gz
As a first step we subset to the ATAC modality.
atac = mdata.mod["atac"]
Although, we are now not working with RNA-Seq, it is possible to use some of scanpy’s preprocessing functions which can also be used on ATAC data. This is possible due to similar distribution and quality issues of both modalities. The only thing to bear in mind here that a gene would mean a peak in the context of the AnnData object with ATAC-seq data. Afterwards, ATAC specific preprocessing can be conducted with the ATAC module of muon.
Let us start with some quality control by filtering out cells with too few peaks and peaks detected in too few cells. For now, we will filter out cells that do not pass QC.
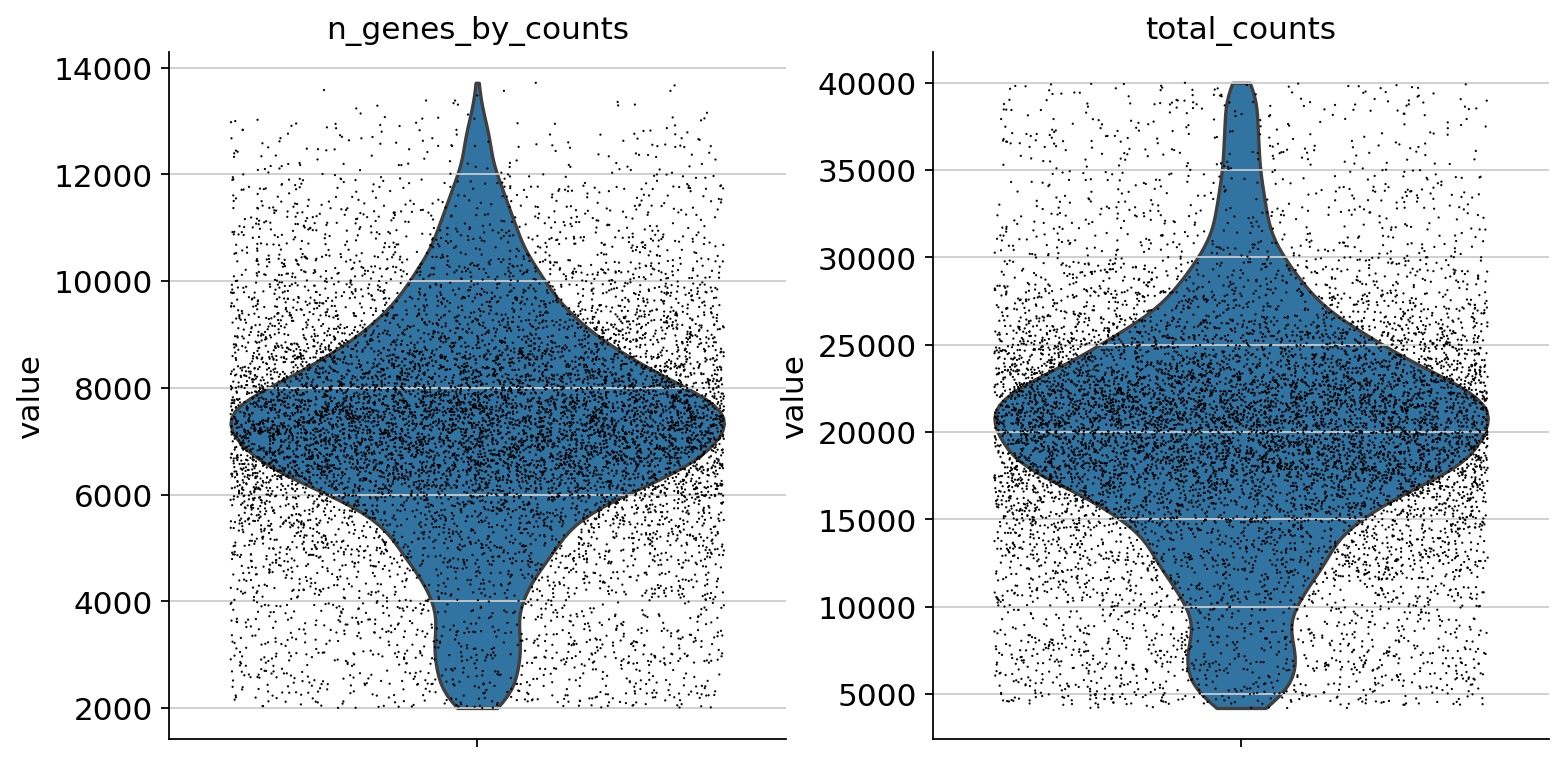
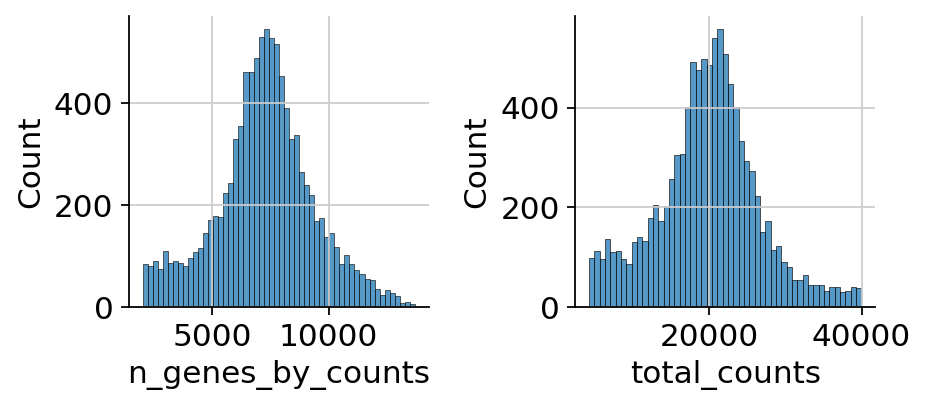
sc.pp.calculate_qc_metrics(atac, percent_top=None, log1p=False, inplace=True)
sc.pl.violin(atac, ["total_counts", "n_genes_by_counts"], jitter=0.4, multi_panel=True)
/home/zeth/miniconda3/envs/bp2/lib/python3.9/site-packages/anndata/_core/anndata.py:1228: FutureWarning: The `inplace` parameter in pandas.Categorical.reorder_categories is deprecated and will be removed in a future version. Reordering categories will always return a new Categorical object.
c.reorder_categories(natsorted(c.categories), inplace=True)
... storing 'feature_types' as categorical
/home/zeth/miniconda3/envs/bp2/lib/python3.9/site-packages/anndata/_core/anndata.py:1228: FutureWarning: The `inplace` parameter in pandas.Categorical.reorder_categories is deprecated and will be removed in a future version. Reordering categories will always return a new Categorical object.
c.reorder_categories(natsorted(c.categories), inplace=True)
... storing 'genome' as categorical
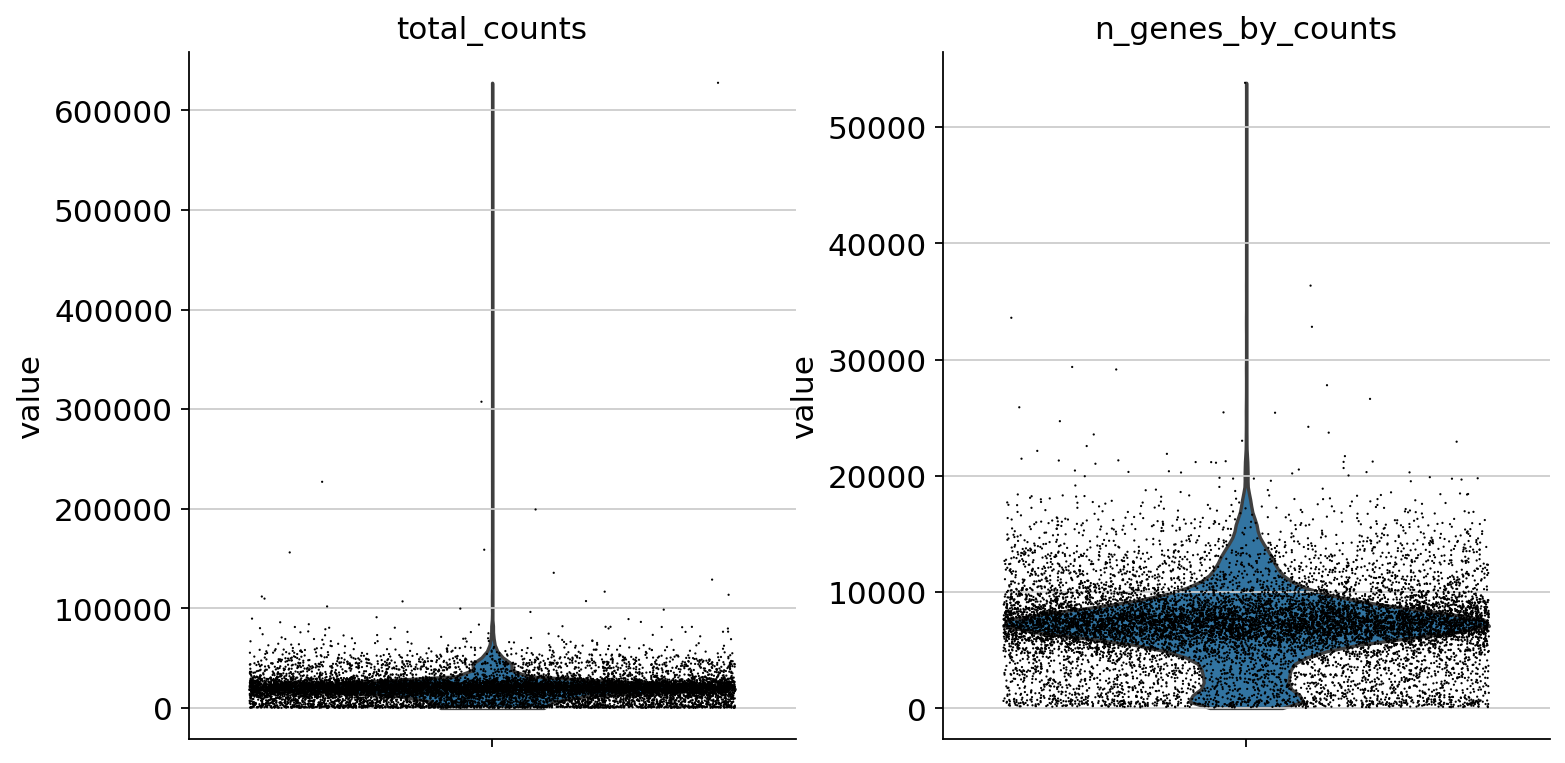
Filter peaks whose expression is not detected.
mu.pp.filter_var(atac, "n_cells_by_counts", lambda x: x >= 10)
# This is analogous to
# sc.pp.filter_genes(rna, min_cells=10)
# but does in-place filtering and avoids copying the object
We also filter the cells.
mu.pp.filter_obs(atac, "n_genes_by_counts", lambda x: (x >= 2000) & (x <= 15000))
# This is analogous to
# sc.pp.filter_cells(atac, max_genes=15000)
# sc.pp.filter_cells(atac, min_genes=2000)
# but does in-place filtering avoiding copying the object
mu.pp.filter_obs(atac, "total_counts", lambda x: (x >= 4000) & (x <= 40000))
muon also provides histograms which allows for a different view on the metrics.
Now that we rudimentary filtered out cells with too few peaks and peaks detected in too few cells, we can start with ATAC specific quality control with muon. muon has modality specific preprocessing functions in corresponding modules. We import the ATAC module to access the ATAC specific preprocessing functions.
from muon import atac as ac
# Perform rudimentary quality control with muon's ATAC module
atac.obs["NS"] = 1
ac.tl.nucleosome_signal(atac, n=1e6)
ac.tl.get_gene_annotation_from_rna(mdata["rna"]).head(3)
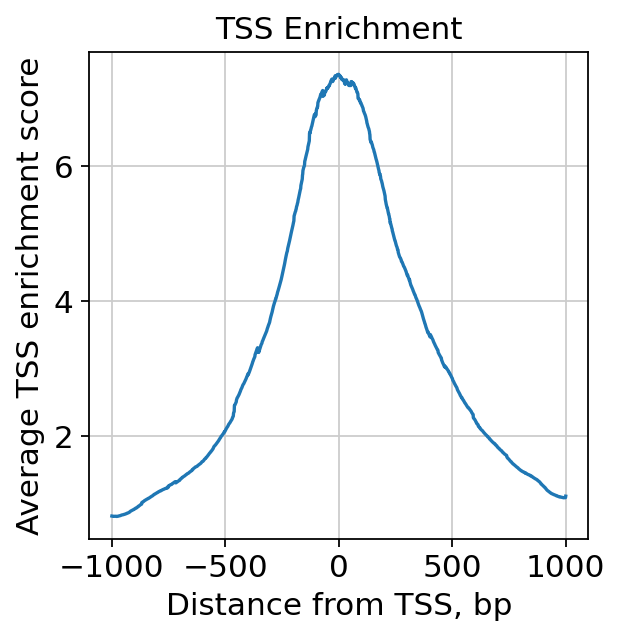
tss = ac.tl.tss_enrichment(mdata, n_tss=1000)
ac.pl.tss_enrichment(tss)
# Save original counts, normalize data and select highly variable genes with scanpy
atac.layers["counts"] = atac.X
sc.pp.normalize_per_cell(atac, counts_per_cell_after=1e4)
sc.pp.log1p(atac)
sc.pp.highly_variable_genes(atac, min_mean=0.05, max_mean=1.5, min_disp=0.5)
atac.raw = atac
Although PCA is also commonly used for ATAC data, latent semantic indexing (LSI) is another popular option. It is implemented in muon’s ATAC module.
ac.tl.lsi(atac)
sc.pp.neighbors(atac, use_rep="X_lsi", n_neighbors=10, n_pcs=30)
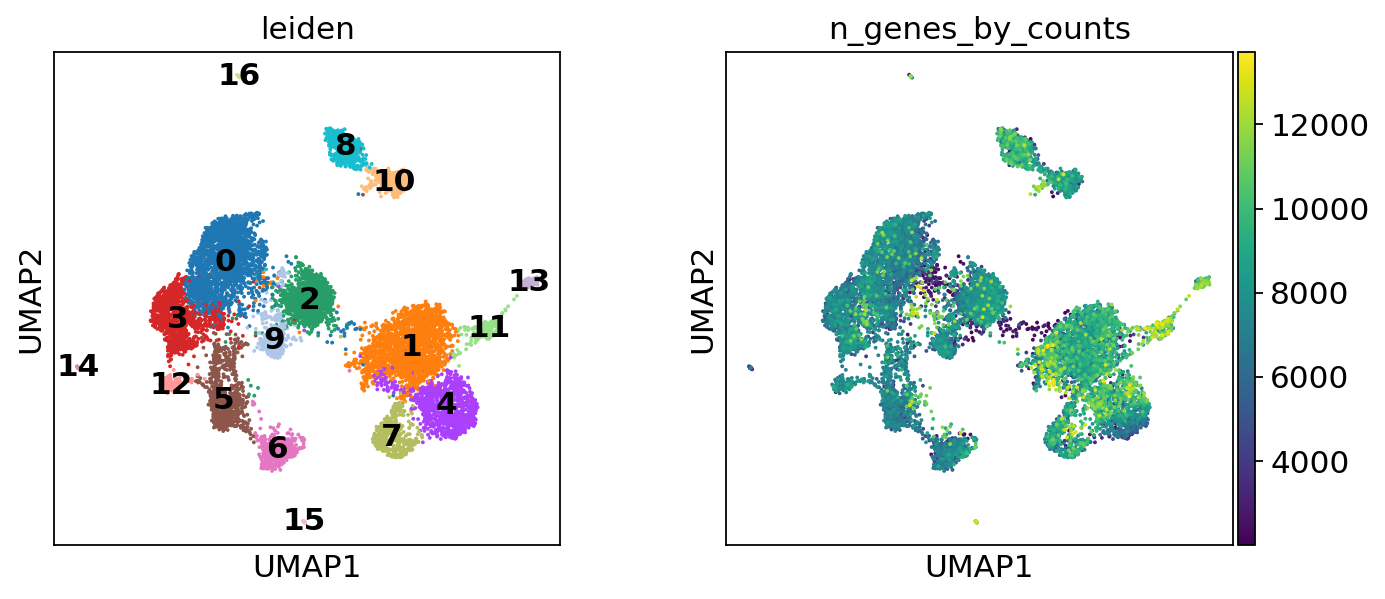
sc.tl.leiden(atac, resolution=0.5)
sc.tl.umap(atac, spread=1.5, min_dist=0.5, random_state=20)
sc.pl.umap(atac, color=["leiden", "n_genes_by_counts"], legend_loc="on data")
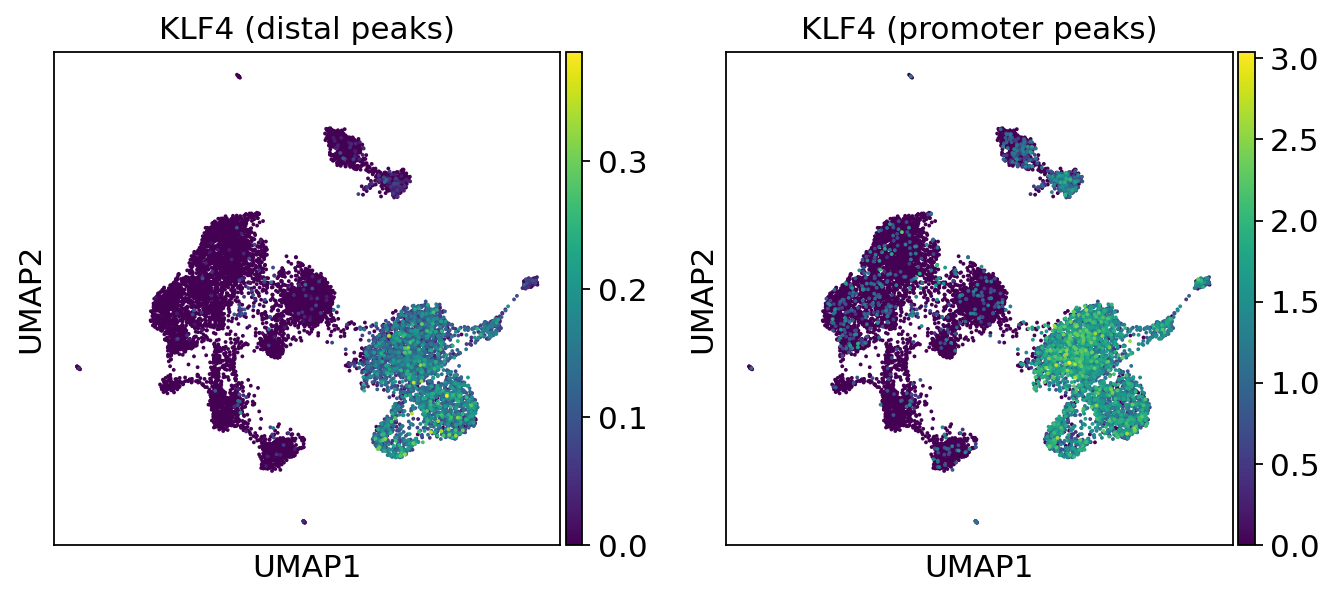
We can use the functionality of the ATAC module in muon to color plots by cut values in peaks corresponding to a certain gene.
For more details on all available functions of muon, please read the muon API reference at https://muon.readthedocs.io/en/latest/api/index.html and the muon tutorials at https://muon-tutorials.readthedocs.io/en/latest/.
4.6. References#
Danila Bredikhin, Ilia Kats, and Oliver Stegle. Muon: multimodal omics analysis framework. Genome Biology, 23(1):42, Feb 2022. URL: https://doi.org/10.1186/s13059-021-02577-8, doi:10.1186/s13059-021-02577-8.
Yuhan Hao, Stephanie Hao, Erica Andersen-Nissen, William M. Mauck III, Shiwei Zheng, Andrew Butler, Maddie J. Lee, Aaron J. Wilk, Charlotte Darby, Michael Zagar, Paul Hoffman, Marlon Stoeckius, Efthymia Papalexi, Eleni P. Mimitou, Jaison Jain, Avi Srivastava, Tim Stuart, Lamar B. Fleming, Bertrand Yeung, Angela J. Rogers, Juliana M. McElrath, Catherine A. Blish, Raphael Gottardo, Peter Smibert, and Rahul Satija. Integrated analysis of multimodal single-cell data. Cell, 2021. URL: https://doi.org/10.1016/j.cell.2021.04.048, doi:10.1016/j.cell.2021.04.048.
Wolfgang Huber, Vincent J. Carey, Robert Gentleman, Simon Anders, Marc Carlson, Benilton S. Carvalho, Hector Corrada Bravo, Sean Davis, Laurent Gatto, Thomas Girke, Raphael Gottardo, Florian Hahne, Kasper D. Hansen, Rafael A. Irizarry, Michael Lawrence, Michael I. Love, James MacDonald, Valerie Obenchain, Andrzej K. Oleś, Hervé Pagès, Alejandro Reyes, Paul Shannon, Gordon K. Smyth, Dan Tenenbaum, Levi Waldron, and Martin Morgan. Orchestrating high-throughput genomic analysis with bioconductor. Nature Methods, 12(2):115–121, Feb 2015. URL: https://doi.org/10.1038/nmeth.3252, doi:10.1038/nmeth.3252.
Project Jupyter. Jupyter. https://jupyter.org/, 2022. Accessed: 2022-04-21.
Combiz Khozoie, Nurun Fancy, Mahdi M. Marjaneh, Alan E. Murphy, Paul M. Matthews, and Nathan Skene. Scflow: a scalable and reproducible analysis pipeline for single-cell term`rna` sequencing data. bioRxiv, 2021. URL: https://www.biorxiv.org/content/early/2021/08/19/2021.08.16.456499.1, arXiv:https://www.biorxiv.org/content/early/2021/08/19/2021.08.16.456499.1.full.pdf, doi:10.1101/2021.08.16.456499.
Giovanni Palla, Hannah Spitzer, Michal Klein, David Fischer, Anna Christina Schaar, Louis Benedikt Kuemmerle, Sergei Rybakov, Ignacio L. Ibarra, Olle Holmberg, Isaac Virshup, Mohammad Lotfollahi, Sabrina Richter, and Fabian J. Theis. Squidpy: a scalable framework for spatial omics analysis. Nature Methods, 19(2):171–178, Feb 2022. URL: https://doi.org/10.1038/s41592-021-01358-2, doi:10.1038/s41592-021-01358-2.
scverse. Scverse. https://scverse.org, 2022. Accessed: 2022-04-21.
scverse mudata. Mudata quickstart. https://mudata.readthedocs.io/en/latest/notebooks/quickstart_mudata.html, 2022. Accessed: 2022-04-21.
scverse mudata. Mudata quickstart. https://mudata.readthedocs.io/en/latest/io/mudata.html, 2022. Accessed: 2022-04-21.
scverse scanpy. Scanpy api. https://scanpy.readthedocs.io/en/stable/api.html#, 2022. Accessed: 2022-04-21.
Gregor Sturm, Tamas Szabo, Georgios Fotakis, Marlene Haider, Dietmar Rieder, Zlatko Trajanoski, and Francesca Finotello. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics, 36(18):4817–4818, 07 2020. URL: https://doi.org/10.1093/bioinformatics/btaa611, arXiv:https://academic.oup.com/bioinformatics/article-pdf/36/18/4817/34560298/btaa611.pdf, doi:10.1093/bioinformatics/btaa611.
Isaac Virshup, Sergei Rybakov, Fabian J. Theis, Philipp Angerer, and F. Alexander Wolf. Anndata: annotated data. bioRxiv, 2021. URL: https://www.biorxiv.org/content/early/2021/12/19/2021.12.16.473007, arXiv:https://www.biorxiv.org/content/early/2021/12/19/2021.12.16.473007.full.pdf, doi:10.1101/2021.12.16.473007.
F. Alexander Wolf, Philipp Angerer, and Fabian J. Theis. Scanpy: large-scale single-cell gene expression data analysis. Genome Biology, 19(1):15, Feb 2018. URL: https://doi.org/10.1186/s13059-017-1382-0, doi:10.1186/s13059-017-1382-0.
4.7. Contributors#
We gratefully acknowledge the contributions of:
4.7.2. Reviewers#
Isaac Virshup